Tutorial
Updated:2025-08-28 (Latest)
Below step-by-step guideline will walk you through everything you need to participate in the Medprompt Hackathon 2025 on Kaggle. Whether you’re new to data science competitions or an experienced participant, this process is designed to be simple and accessible:
No programming required: You will not need to write code or have any programming experience. Your main task is to edit and optimize a single prompt in a provided Jupyter notebook.
No high-end hardware needed: All work takes place on the Kaggle platform using their cloud servers. You don’t need a powerful computer or special setup—just an internet connection and a Kaggle account.
No model adjustments necessary: The language model and all settings are fixed. For final evaluation, only your edited prompt will be used. Please focus solely on prompt optimization.
In this guideline, you will learn how to:
Join the competition and access all required materials,
Download and import the provided Jupyter notebook,
Locate and edit the prompt section to test your ideas,
Run the notebook to generate predictions and outputs,
Adjust the number of cases you process,
Submit your results for evaluation—directly through Kaggle.
Detailed Guidelines
These guidelines are intended solely to help you set up your prompt environment for the Medprompt Hackathon 2025 on Kaggle. All images, screenshots, and sample texts are for demonstration purposes only. For the final version of Competition details, descriptions, and titles, please refer to the official competition websites on Kaggle.
1) Join the Competition
Use the invitation link to join the competition first.
2) Download the Jupyter Notebook
Go to the “Data” tab on the competition page.
Scroll down to find the list of available notebooks. Each notebook specifies the LLM it uses in its filename or description. Download the notebook that matches the LLM you want to use. You can download just the notebook or all competition files if needed.
Models available:
• DeepSeek R1:14B
• Gemma3:12B
• Gemma3:27B
• Qwen3:14B
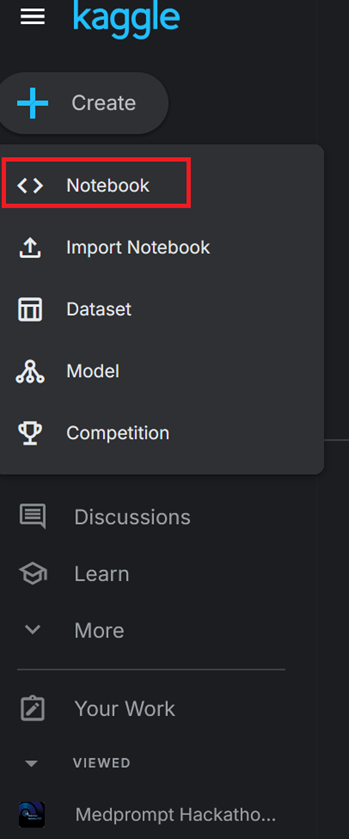
3)Open the side panel on the Kaggle website and select “Create > Notebook”
4) On the notebook page, go to File > Import Notebook (top left corner). Import the Jupyter notebook you downloaded earlier.
5) Prompt adjustment
Scroll down to code block #6 and locate the prompt section to make any adjustments as needed.
{case_text} is a placeholder in your prompt that will be replaced by the actual case report text for each input.
<DIAGNOSIS>...</DIAGNOSIS> are special tags you should use if you want to mark your diagnosis in the model’s output.
The system looks for text between these tags to extract your answer automatically.
Example:
If your answer is:
<DIAGNOSIS>Pneumonia</DIAGNOSIS>
the system will extract “Pneumonia” only as your diagnosis.
6) Run the Notebook
For the initial setup, select “Run All” to execute all cells and set up the environment for running the LLM. If the session remains active and you only make changes to the prompts, you can run only code blocks #6 and #7.
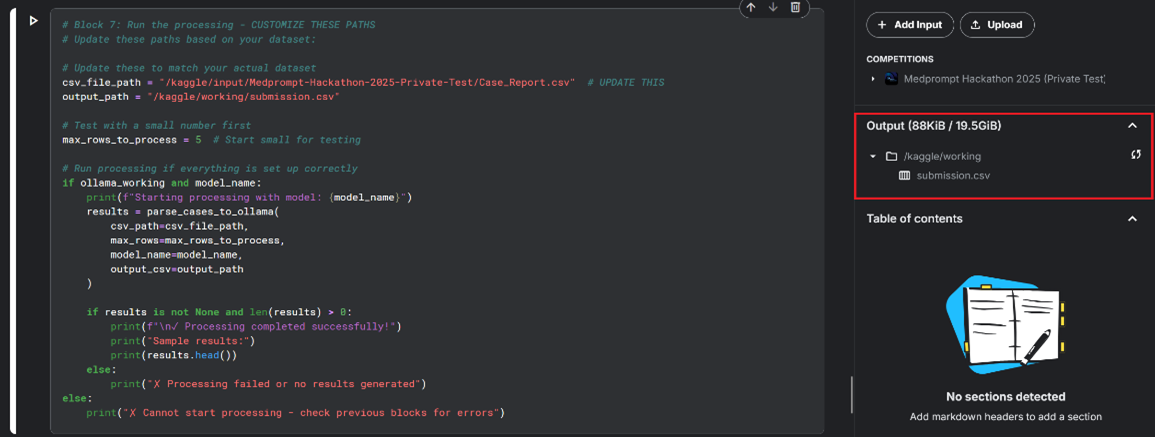
7) Check your results in the “Output” tab on the right side panel.
8) Adjust the Number of Rows
By default, the maximum number of rows processed is 5. Update the value in code block #7 to modify. (Row numbers for the complete file are 200).
9) Click the Submit button in the “Submit to competition” tab on the right side panel to evaluate your submission. You do not need to download the submission file manually—just click submit. (Note: need the full 200 rows to evaluate)
Your prompt will be automatically included in the submission file along with your generated diagnosis. The second row will automatically include information about the LLM (Large Language Model) you used.
Note:
Kaggle provides a weekly quota for accelerator usage. To conserve your quota, it is recommended to turn off your session when not in use. However, if you turn off the session, you will need to re-run all code blocks to re-establish the environment before running the LLM again.
If you are using the 27B model, please be aware that it will take significantly more time to process full case reports compared to smaller models.